A new study published in *Genome Biology* on June 27, 2025, sheds light on the intricate 3D structure of the human genome and its significant role in regulating gene activity. Researchers from **Sanford Burnham Prebys** and their colleagues in **Hong Kong** have demonstrated that the three-dimensional arrangement of chromatin—complexed DNA and proteins—can influence gene regulation, offering insight into potential links to various diseases, including cancers.
Traditionally, biology education presents the human genome in a one-dimensional format, focusing on how sequences of DNA form genes. While this linear perspective is essential for understanding genetic basics, it fails to capture the complexity of the genome’s spatial configuration. Inside the nucleus of each cell, approximately six feet of DNA is meticulously coiled around proteins known as histones, forming a compact structure called chromatin. This arrangement features numerous loops and clusters that, while appearing chaotic, facilitate interactions between certain genomic regions while isolating others.
The research team identified that nearly 12% of genomic regions in breast cancer cells exhibit structural abnormalities within their chromatin. Such issues are also linked to conditions like T-cell acute lymphoblastic leukemia, underscoring the importance of understanding chromatin’s architecture.
Exploring the Role of 3D Structure in Gene Regulation
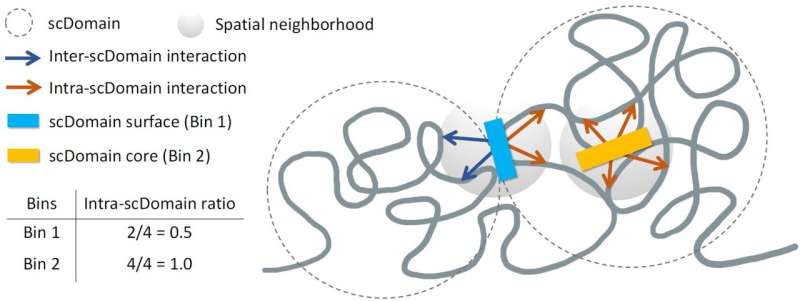
Dr. **Kelly Yichen Li**, a postdoctoral associate at Sanford Burnham Prebys and the study’s lead author, emphasized the significance of their findings: “We know that many regions of the genome tend to form what are known as topologically associating domains or TADs. Parts of the genome within these domains can interact more frequently with each other, while they tend to be isolated from the region outside this domain.”
In their investigation, the researchers employed advanced imaging techniques to spatially map chromatin in individual cells. They observed that TAD-like regions often took on a globular shape, resembling the varied forms of potatoes. This unique morphology suggests that the 3D structure of these regions may play a crucial role in regulating nearby gene activity.
Dr. **Yuk-Lap (Kevin) Yip**, a professor and interim director of the Center for Data Sciences at Sanford Burnham Prebys, noted, “If you picture these clumps of chromatin fiber being roughly in the shape of a potato, we predicted that regions of the genome closer to the surface are more active due to exposure to nearby biochemical signals in the cell nucleus.”
The researchers likened the protective function of a potato’s skin to the chromatin’s core structure, positing that genes located deeper within these clumps may experience reduced accessibility to signals that promote gene expression.
Innovative Methods and Future Directions
To validate their hypotheses, the team developed a new metric to quantify the “coreness” of genomic regions within chromatin domains. Dr. Li explained, “This measure allowed us to define the surface and core, and we went on to show that surface regions are more active than core regions.” This approach opens avenues for further research, as the data generated can be applied to investigate how the ‘coreness’ of chromatin relates to gene activity and disease across various cell types.
Looking forward, Dr. Yip and Dr. Li plan to collaborate with Dr. **Pier Lorenzo Puri**, a medical doctor, to delve deeper into the implications of chromatin structure on muscle stem cell development and the progression of muscular dystrophy. Their work represents a significant step in understanding the complex relationship between genomic architecture and health, highlighting the potential for new therapeutic strategies targeting gene regulation.
For further details, the full study is available in *Genome Biology*, authored by Kelly Yichen Li et al.






