Researchers at the University of Tokyo, alongside other institutions, have made an unexpected discovery in human saliva: giant DNA elements known as Inocles. This finding sheds light on the adaptability of oral bacteria to their environment and opens new avenues for understanding oral health and disease.
Revolutionizing Our Understanding of the Oral Microbiome
Despite significant advancements in medical science, the human microbiome—especially the oral microbiome—continues to reveal surprises. In recent years, research has highlighted various previously unknown aspects of human biology. The latest breakthrough stems from a study led by Project Research Associate Yuya Kiguchi and his team, who analyzed a large collection of saliva samples from the Yutaka Suzuki Lab at the Graduate School of Frontier Sciences.
Inspired by discoveries of extraneous DNA in soil microbiomes, the researchers aimed to identify similar elements in human saliva. Kiguchi noted, “We know there are a lot of different kinds of bacteria in the oral microbiome, but many of their functions and means of carrying out those functions are still unknown.” This exploration led to the discovery of Inocles, which are a type of extrachromosomal DNA—DNA that exists outside the main genetic structure of bacteria.
Advanced Techniques Reveal Hidden Genetic Elements
Detecting Inocles posed significant challenges, as traditional sequencing methods fragment genetic data. To address this, the research team employed advanced long-read sequencing techniques. A pivotal moment in the research came from co-first author Nagisa Hamamoto, who developed a method called preNuc. This technique effectively removed human DNA from saliva samples, enhancing the sequencing quality of other DNA.
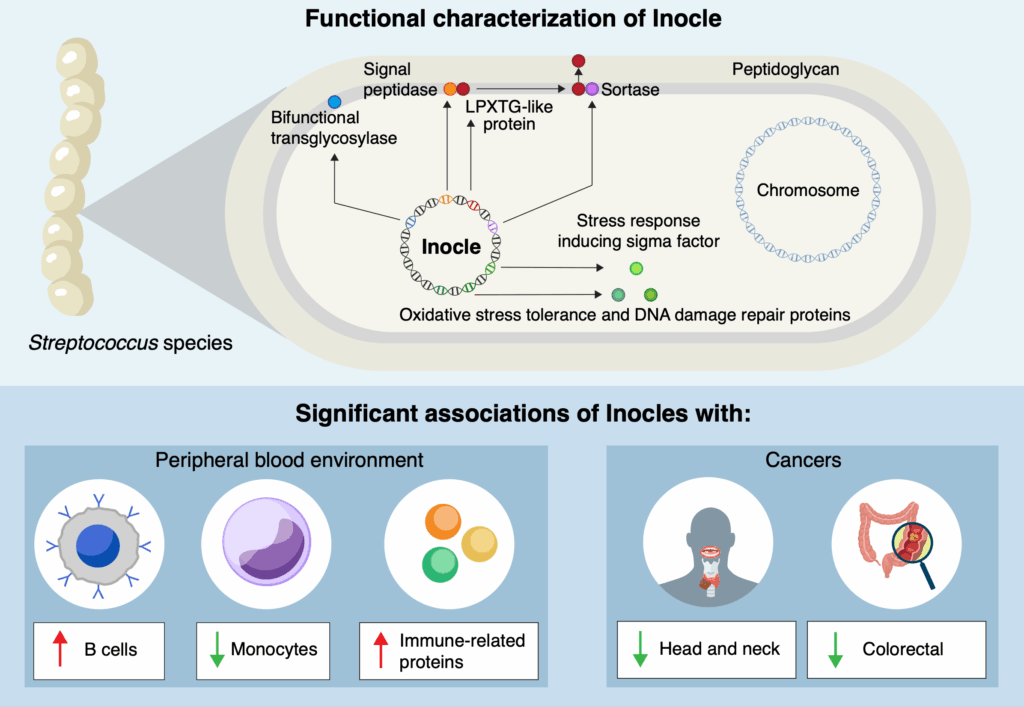
For the first time, the researchers successfully assembled complete Inocle genomes, which were found to be hosted by the bacteria Streptococcus salivarius. Kiguchi explained, “The average genome size of Inocle is 350 kilobase pairs, making it one of the largest extrachromosomal genetic elements in the human microbiome.” In contrast, plasmids, another form of extrachromosomal DNA, typically measure only a few tens of kilobase pairs.
This substantial size equips Inocles with various genes, including those for oxidative stress resistance and DNA damage repair, which could be vital for adapting to environmental challenges.
The research team plans to develop stable methods for culturing bacteria that contain Inocles. This will enable them to further investigate the functionality of these DNA elements, their potential to spread between individuals, and their influence on oral health conditions such as cavities and gum disease.
As many Inocle genes remain uncharacterized, researchers will combine laboratory experiments with computational simulations, including the use of AlphaFold to predict and model the roles of Inocles.
Kiguchi emphasized the significance of their findings, stating, “Given the range of the human population represented by the saliva samples, we believe 74% of all humans may possess Inocles.” He noted that despite extensive studies on the oral microbiome, these elements had remained hidden due to technological limitations.
Now that Inocles have been identified, researchers can explore their impact on the relationship between humans and their resident microbes, particularly concerning oral health. There is even preliminary evidence suggesting that Inocles might serve as markers for serious diseases, including cancer.
This groundbreaking research not only enhances our understanding of the oral microbiome but also has the potential to inform future health strategies and interventions aimed at improving oral and overall health.






